| oligos_5-8nt_m1_shift0 (oligos_5-8nt_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_5-8nt_m1; m=0 (reference); ncol1=11; shift=0; ncol=13; wwCTCGCAwrw--
; Alignment reference
a 11 11 0 4 0 0 0 33 9 9 9 0 0
c 7 8 33 0 34 0 25 0 5 1 7 0 0
g 6 4 1 4 0 33 3 1 3 17 6 0 0
t 10 11 0 26 0 1 6 0 17 7 12 0 0
|
| TOE1_TOE1_ArabidopsisPBM_shift0 (TOE1:TOE1:ArabidopsisPBM) |
 |
0.701 |
0.637 |
5.885 |
0.885 |
0.869 |
6 |
1 |
2 |
2 |
4 |
3.000 |
2 |
; oligos_5-8nt_m1 versus TOE1_TOE1_ArabidopsisPBM (TOE1:TOE1:ArabidopsisPBM); m=2/6; ncol2=10; w=10; offset=0; strand=D; shift=0; score= 3; WCCTCGTACy---
; cor=0.701; Ncor=0.637; logoDP=5.885; NsEucl=0.885; NSW=0.869; rcor=6; rNcor=1; rlogoDP=2; rNsEucl=2; rNSW=4; rank_mean=3.000; match_rank=2
a 69 1 13 0 5 1 1 96 5 23 0 0 0
c 2 88 84 1 93 1 0 2 80 31 0 0 0
g 2 0 1 1 2 97 19 0 5 21 0 0 0
t 27 11 2 98 0 1 80 2 10 25 0 0 0
|
| TOE1_TOE1_2_ArabidopsisPBM_rc_shift3 (TOE1:TOE1_2:ArabidopsisPBM_rc) |
 |
0.737 |
0.454 |
0.733 |
0.883 |
0.891 |
3 |
3 |
5 |
3 |
2 |
3.200 |
3 |
; oligos_5-8nt_m1 versus TOE1_TOE1_2_ArabidopsisPBM_rc (TOE1:TOE1_2:ArabidopsisPBM_rc); m=3/6; ncol2=10; w=8; offset=3; strand=R; shift=3; score= 3.2; ---TCsYAgGTTy
; cor=0.737; Ncor=0.454; logoDP=0.733; NsEucl=0.883; NSW=0.891; rcor=3; rNcor=3; rlogoDP=5; rNsEucl=3; rNSW=2; rank_mean=3.200; match_rank=3
a 0 0 0 6 3 4 4 92 2 3 19 18 21
c 0 0 0 1 93 25 26 1 21 2 1 4 32
g 0 0 0 3 1 63 2 3 64 92 1 3 5
t 0 0 0 90 3 8 68 4 13 3 79 75 42
|
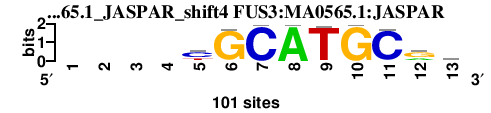
| FUS3_MA0565.1_JASPAR_shift4 (FUS3:MA0565.1:JASPAR) |
 |
0.772 |
0.416 |
4.233 |
0.865 |
0.872 |
2 |
4 |
3 |
4 |
3 |
3.200 |
4 |
; oligos_5-8nt_m1 versus FUS3_MA0565.1_JASPAR (FUS3:MA0565.1:JASPAR); m=4/6; ncol2=9; w=7; offset=4; strand=D; shift=4; score= 3.2; ----yGCATGCrs
; cor=0.772; Ncor=0.416; logoDP=4.233; NsEucl=0.865; NSW=0.872; rcor=2; rNcor=4; rlogoDP=3; rNsEucl=4; rNSW=3; rank_mean=3.200; match_rank=4
a 0 0 0 0 8 1 1 99 1 0 1 29 15
c 0 0 0 0 61 2 99 0 0 0 97 3 44
g 0 0 0 0 4 97 0 0 1 99 2 59 25
t 0 0 0 0 28 1 0 1 99 1 1 9 16
|
| At3g54990_MA0553.1_JASPAR_shift1 (At3g54990:MA0553.1:JASPAR) |
 |
0.710 |
0.516 |
6.601 |
0.860 |
0.842 |
5 |
2 |
1 |
5 |
6 |
3.800 |
5 |
; oligos_5-8nt_m1 versus At3g54990_MA0553.1_JASPAR (At3g54990:MA0553.1:JASPAR); m=5/6; ncol2=8; w=8; offset=1; strand=D; shift=1; score= 3.8; -YCTCGTAC----
; cor=0.710; Ncor=0.516; logoDP=6.601; NsEucl=0.860; NSW=0.842; rcor=5; rNcor=2; rlogoDP=1; rNsEucl=5; rNSW=6; rank_mean=3.800; match_rank=5
a 0 0 0 0 0 0 0 135 0 0 0 0 0
c 0 105 119 0 147 0 0 12 147 0 0 0 0
g 0 0 28 0 0 147 25 0 0 0 0 0 0
t 0 42 0 147 0 0 122 0 0 0 0 0 0
|




